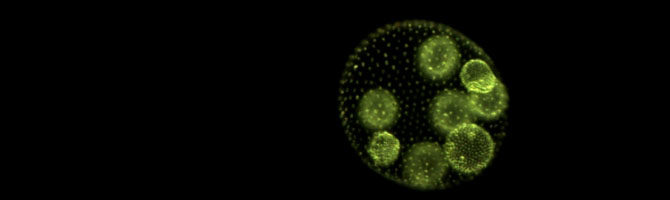
Our paper “De novo origins of multicellularity in response to predation” is #2 on Scientific Reports Top 100 list of most downloaded articles of 2019, with 164k accesses.
Author Archives: Matthew Herron
New preprint: What are the Major Transitions?
EDIT April 1, 2020: updated link and slightly revised abstract to the new version
I have posted a new preprint to the PhilSci Archive:
The ‘Major Transitions in Evolution’ (MTE) framework has emerged as the dominant paradigm for understanding the origins of life’s hierarchical organization, but it has been criticized on the grounds that it lacks theoretical unity, that is, that the events that make up the category do not constitute a natural kind. I agree with this criticism, and I argue that the best response is to modify the category so that it does approximate a natural kind. Specifically, I recommend defining major transitions as all those, and only those, events and processes that result in the emergence of a new level of selection. Two sorts of changes will be required to achieve this. First, events and processes that do not meet this criterion, such as the origins of the genetic code and of human language, should be excluded. Second, events and processes that do meet the criterion, but which have generally been neglected, should be included. These changes would have the dual benefits of making MTEs a philosophically coherent category and of increasing the sample size on which we may infer trends and general principles that may apply to all MTEs.
New position
I have started a stint as a rotating program officer at the National Science Foundation (Division of Environmental Biology, Evolutionary Processes Cluster). While I serve at NSF, I’ll be on a leave of absence from Georgia Tech, but I will visit periodically while my lab continues to operate.
New article in The Science Breaker
Postdoc Kimberly Chen has published a lay summary of our recent Scientific Reports paper, in which we showed that predation can drive the evolution of multicellularity in the green alga Chlamydomonas:
Multicellular life is one of the most astonishing wonders on Earth, but why and how does it arise in the first place, and at what cost? To help answer these questions, we exposed single-celled algae to predators and watched them evolve into multicellular life. Within a year, they had formed groups of cells to avoid being eaten – but at a price.
Chen, I-C. K. & M. D. Herron. 2019. Predators drive the evolution of multicellularity. The Science Breaker 257. doi: 10.25250/thescbr.brk257
NSF Project Outcomes Report

The Project Outcomes Report for our recently ended NSF grant (known variously as DEB-1723293, DEB-1457701, and DEB-1456652) is now available on grants.gov:
Life comes in two forms: single-celled (organisms made up of only one cell) and multi-celled (organisms made up of many cells, alike or different). Multicellular life has evolved from unicellular ancestors many times across the tree of life, and the resulting radiations have transformed nearly every ecosystem on Earth. Ancestors of animals, plants, fungi, several groups of seaweeds, and filamentous bacteria underwent the transition from single- to multi-celled life in the deep past. While each of these origins is a replicate experiment with the potential to inform our understanding of how and why multicellular life evolved, the window through which we see these ancient events is blurry. Extinctions, subsequent evolution, and a spotty fossil record obscure our view. Experimental evolution enables us to time-travel, making it possible to clearly observe the evolution of multicellularity as it occurs in the lab. This project integrated experimental, bioinformatic, theoretical, and comparative approaches to understand how multicellularity and related traits have evolved, and how they can evolve.
Fungi are old

Fungi are weird

Volvox 2019 poster

New paper in Evolution
A new paper describing the results of a yeast evolution experiment has been published in Evolution. Jordan Gulli exposed nascent multicellular “snowflake yeast” to an environment in which solitary multicellular clusters experienced low survival. In response, snowflake yeast evolved to form cooperative groups composed of thousands of multicellular clusters.

Predation project on Common Descent podcast
The Common Descent podcast spent a few minutes talking about the new Scientific Reports paper (~18:25-24:30):